Predict Distribution Functions Across Geographic Space
Source:R/predict_functions.r
predict_functions.RdPredicts constrained, univariate distribution functions across the geographic space supported by raster layers. For a given continuous variable used to create a classification unit, this function first calculates a user-defined distribution function for that variable using only observations selected from within the classification unit. In this way, the distribution function is univariate and constrained. Subsequently, a locally-estimated scatterplot smoothing (LOESS) or a generalized additive model (GAM) is fitted. This model is fitted using the variable’s observations as explanatory values and the values from the distribution function as the response values. Finally, the fitted model is predicted on the complete geographic space supported by the raster layer of the given variable. This process is iterated for all of the continuous variables and classification units. Each resulting layer can be thought of as a landscape correspondence measurement between an XY location in geographic space and the landscape configuration represented by a given classification unit in terms of a specific variable. The following distribution functions are currently supported: the probability density function (PDF), the empirical cumulative density function (ECDF), and the inverse of the empirical cumulative density function (iECDF). Please refer to Details for more information about how each distribution function is calculated. Also, see details on parallel processing.
Usage
predict_functions(
cuvar.rast,
cu.ind,
cu,
vars,
dif,
hist.type = "regular",
hist.pen = "default",
grid.mult = 1,
kern = "normal",
quant.sep = 0.01,
method = "loess",
span = 0.6,
k = 20,
discrete = TRUE,
to.disk = FALSE,
outdir = ".",
prefix = "",
extension = ".tif",
overwrite = FALSE,
verbose = FALSE,
...
)Arguments
- cuvar.rast
SpatRaster, as in
rast. Multi-layer SpatRaster containing n continuous raster layers (i.e., variables) and one raster layer of classification units with integer cell values (i.e., Numeric identifiers).- cu.ind
Integer. Position (index) of the raster layer of classification units in cuvar.rast.
- cu
Integer. Vector of integer values that correspond to the numeric identifiers of the units in the raster layer of classification units.
- vars
Character. Vector of strings containing the names of the n continuous variables in cuvar.rast. These names have to be sequentially repeated according to the number of classification units (See Examples).
- dif
Character. Vector of strings containing the distribution function to calculate for each continuous variable within each classification unit. The function will match the position of the name of the distribution function with that of the name of the continuous variable in vars.
- hist.type
Character. Type of histogram to calculate. Options are "regular", "irregular" (unequally-sized bins, very slow), and "combined" (the one with greater penalized likelihood is returned). See
histogram. Default: "regular"- hist.pen
Character. Penalty to apply when calculating the histogram (see
histogram). Default: "default"- grid.mult
Numeric. Multiplying factor to increase/decrease the size of the "optimal" grid size for the Kernel Density Estimate (KDE). Default: 1
- kern
Character. Type of kernel to use for the KDE. Default: "normal"
- quant.sep
Numeric. Spacing between quantiles for the calculation of the ECDF and iECDF. Quantiles are in the range of 0-1 thus spacing must be a decimal. Default: 0.01
- method
Character. Model to fit. Current options are "loess" for locally-estimated scatterplot smoothing (see
loess), and "gam" for generalized additive model with support for large datasets (seebam). Default: "loess"- span
Numeric. If method = "loess", degree of smoothing for LOESS fitting. Default: 0.6
- k
Numeric. If method = "gam", Number of knots for the cubic regression splines. Default: 20
- discrete
Boolean. If method = "gam", discretize variables for storage and efficiency reasons? Can reduce processing time significantly. Default: TRUE
- to.disk
Boolean. Write the output raster layers of predicted distribution function to disk? See details an example about parallel processing. Default: FALSE
- outdir
Character. If to.disk = TRUE, string specifying the path for the output raster layers of predicted distribution function. Default: "."
- prefix
Character. If to.disk = TRUE, string specifying a prefix for the file names of the output raster layers of predicted distribution function. Default: ""
- extension
Character. If to.disk = TRUE, string specifying the extension for the output raster layers of predicted distribution function. Default: ".tif"
- overwrite
Boolean. If to.disk = TRUE, should raster layers in disk and with same name as the output raster layer(s) of predicted distribution function be overwritten? Default: FALSE
- verbose
Boolean. Show warning messages in the console? Default: FALSE
- ...
If to.disk = TRUE, additional arguments as for
writeRaster.
Value
Single-layer or multi-layer SpatRaster with the predicted distribution function for each variable and for each classification unit.
Details
To calculate the PDF, this function uses the binned KDE for observations
drawn from the breaks of a regular/irregular histogram. The "optimal" number
of bins for the histogram is defined by calling the function
histogram (Mildenberger et al., 2019) with the
user-defined penalty hist.pen. Subsequently, the optimal number of
bins is treated as equivalent to the "optimal" grid size for the binned KDE.
The grid size can be adjusted by specifying the multiplying factor
grid.mult. Lastly, the "optimal" bandwidth for the binned KDE is
calculated by applying the direct plugin method of Sheather and Jones
(1991). For the calculation of optimal bandwidth and for the binned KDE, the
package KernSmooth is called. To calculate both the ECDF and the
iECDF, this function calls the ecdf function on
equally-spaced quantiles. The spacing between quantiles can be manually
adjusted via quant.sep. In the case of iECDF, the ECDF is inverted by
applying the formula: iECDF = ((x - max(ECDF)) * -1) + min(ECDF);
where x corresponds to each value of the ECDF.
The "cu", "vars", and "dif" parameters of this function are configured such
that the output table from select_functions can be used
directly as input. (see Examples).
When writing output raster layer to disk, multiple distribution functions can
be predicted in parallel if a parallel backend is registered beforehand with
registerDoParallel. Keep in mind that the function
may require a large amount of memory when using a parallel backend with large
raster layers (i.e., high resolution and/or large spatial coverage).
Some issues have been reported when manually creating cluster objects using
the parallel package. To overcome this issue, a cluster object can be
registered directly through registerDoParallel
without passing it first through makeCluster. See
examples.
References
T. Mildenberger, Y. Rozenholc, and D. Zasada. histogram: Construction of Regular and Irregular Histograms with Different Options for Automatic Choice of Bins, 2019. https://CRAN.R-project.org/package=histogram
S. Sheather and M. Jones. A reliable data-based bandwidth selection method for kernel density estimation. Journal of the Royal Statistical Society. Series B. Methodological, 53:683–690, 1991.
See also
Other Landscape Correspondence Metrics:
select_functions(),
signature,
similarity()
Examples
require(terra)
p <- system.file("exdat", package = "rassta")
# Multi-layer SpatRaster of topographic variables
## 3 continuous variables
ftva <- list.files(path = p, pattern = "^height|^slope|^wetness",
full.names = TRUE
)
tva <- terra::rast(ftva)
# Single-layer SpatRaster of topographic classification units
## Five classification units
ftcu <- list.files(path = p, pattern = "topography.tif", full.names = TRUE)
tcu <- terra::rast(ftcu)
# Add the classification units to the SpatRaster of topographic variables
tcuvars <- c(tcu, tva)
# Data frame with source for "cu", "vars", and "dif"
ftdif <- list.files(path = p, pattern = "topodif.csv", full.names = TRUE)
tdif <- read.csv(ftdif)
# Check structure of source data frame
head(tdif)
#> Class.Unit Variable Dist.Func
#> 1 1 height ECDF
#> 2 1 slope PDF
#> 3 1 wetness PDF
#> 4 2 height PDF
#> 5 2 slope ECDF
#> 6 2 wetness iECDF
# Predict distribution functions
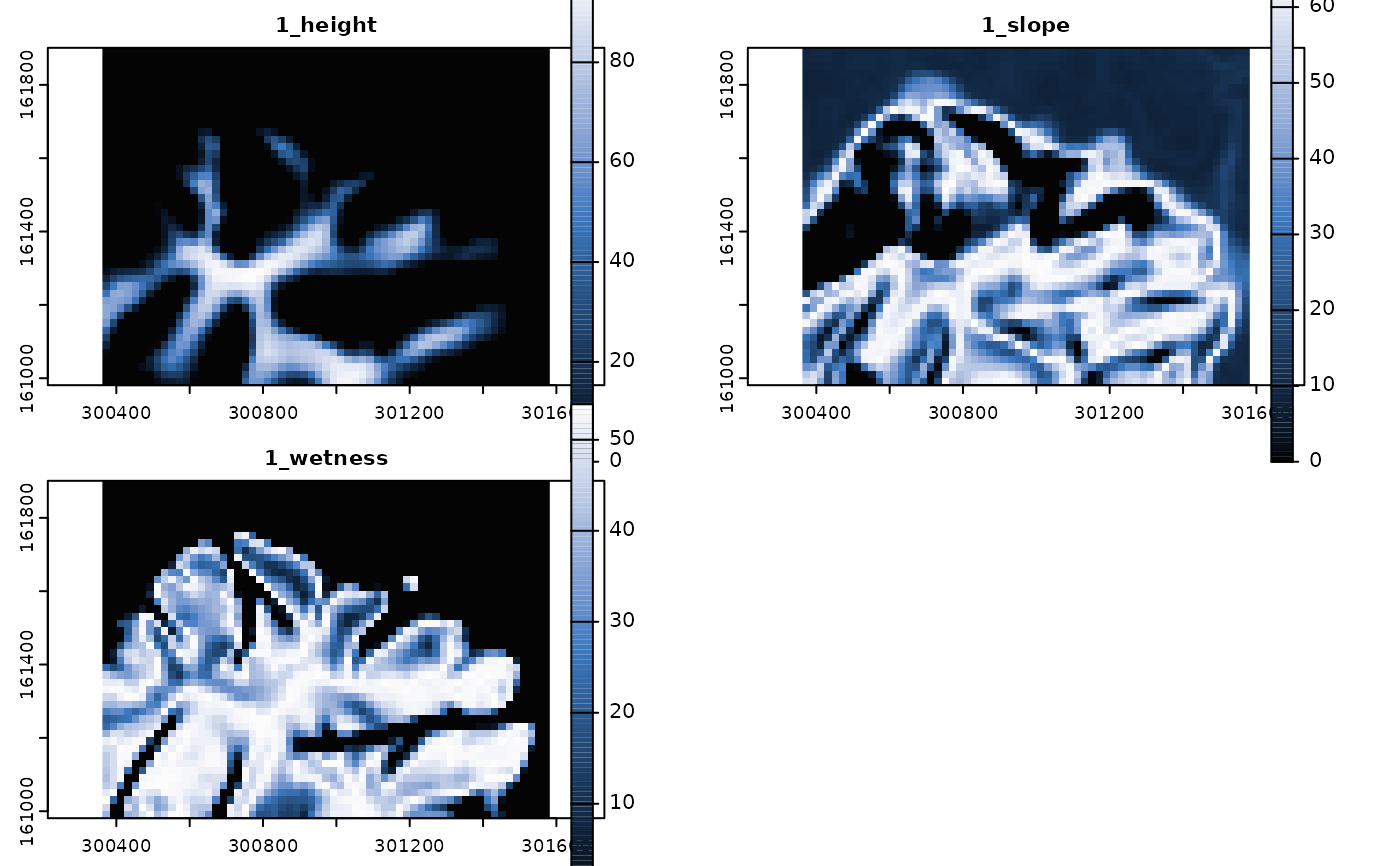
## 1 distribution function per variable and classification unit = 1
tpdif <- predict_functions(cuvar.rast = tcuvars,
cu.ind = 1,
cu = tdif$Class.Unit[1:3],
vars = tdif$Variable[1:3],
dif = tdif$Dist.Func[1:3],
grid.mult = 3,
span = 0.9
)
# Plot predicted distribution functions
if(interactive()){plot(tpdif, col = hcl.colors(100, "Oslo", rev = TRUE))}
 #--------
# Writing results to disk and parallel processing
if(interactive()){
# Directory for temporary files
o <- tempdir()
# Register parallel backend
require(doParallel)
registerDoParallel(4)
# Predict distribution functions
tpdif <- predict_functions(cuvar.rast = tcuvars,
cu.ind = 1,
cu = tdif$Class.Unit[1:3],
vars = tdif$Variable[1:3],
dif = tdif$Dist.Func[1:3],
grid.mult = 3, span = 0.9,
to.disk = TRUE,
outdir = o
)
# Stop cluster
stopImplicitCluster()
# Clean temporary files
file.remove(sources(tpdif))
}
#> [1] TRUE TRUE TRUE
#--------
# Writing results to disk and parallel processing
if(interactive()){
# Directory for temporary files
o <- tempdir()
# Register parallel backend
require(doParallel)
registerDoParallel(4)
# Predict distribution functions
tpdif <- predict_functions(cuvar.rast = tcuvars,
cu.ind = 1,
cu = tdif$Class.Unit[1:3],
vars = tdif$Variable[1:3],
dif = tdif$Dist.Func[1:3],
grid.mult = 3, span = 0.9,
to.disk = TRUE,
outdir = o
)
# Stop cluster
stopImplicitCluster()
# Clean temporary files
file.remove(sources(tpdif))
}
#> [1] TRUE TRUE TRUE